
Pre-historic molecules, recreated
The WSS-funded palaeobiotechnology project in Jena has achieved a major milestone: in an article published in the renowned academic journal Science, Pierre Stallforth, Christina Warinner and their team describe how they recreated microbial agents that lived up to 100,000 years ago.
Bacteria produce all manner of chemical substances—antibiotics, for example, which they need to protect themselves against other bacteria. These substances, called “natural products”, are one of the most important sources for developing new therapeutic drugs. At present, however, researchers are only able to extract the substances from living bacteria.
The palaeobiotechnology project, which received a ten-year grant from the Werner Siemens Foundation in 2020, has now tapped into a new, unusual source of natural products. For the first time, researchers in the teams led by chemist Pierre Stallforth (Friedrich Schiller University Jena and Leibniz Institute for Natural Product Research and Infection Biology) and archaeologist Christina Warinner (Harvard University and the Max Planck Institute for Evolutionary Anthropology) recreated substances produced by bacteria found in the dental plaque of early humans—bacteria that lived 100,000 years ago.

Bacterial genes from dental plaque
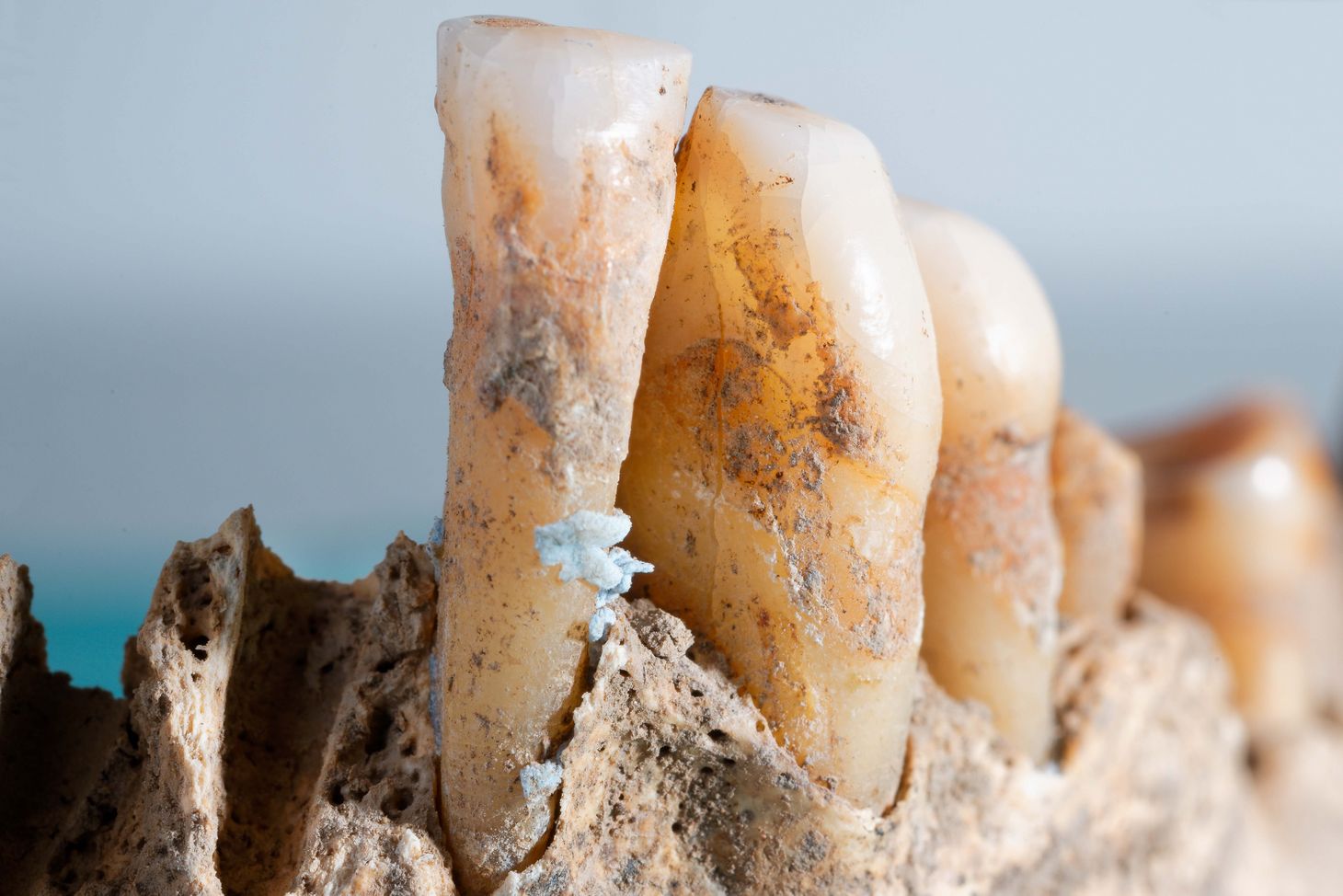
Dental plaque is the only part of the human body that fossilises already during our lifetime, in a process in which living tartar is turned into a graveyard of mineralised bacteria. For the study now published in the top-tier journal Science, the interdisciplinary team led by Warriner and Stallforth examined the dental plaque of Neanderthals, who lived between 40,000 and 100,000 years ago, and of humans, who lived between 150 and 30,000 years ago. They found a wide range of DNA fragments and assembled them—much like pieces of a puzzle—into extremely long DNA sequences with more than 100,000 base pairs.
In doing so, the researchers were able to reconstruct the genomes of numerous bacterial species, many of which are found in the oral flora of present-day humans. However, several reconstructed genomes belonged to more unusual bacterial species that had never been described. One such exotic bacterium was a previously unknown green sulphur bacterium from the genus Chlorobium. “These bacteria aren’t part of the typical oral microbiome,” Pierre Stallforth explains. Nevertheless, genetic material from precisely this bacterium was found in the dental plaque of seven prehistoric humans and Neanderthals.
Although the Chlorobium DNA in the samples was highly damaged—a common problem in ancient genetic samples—the researchers used bioinformatics to remedy the errors and exclude impurities. “The quality of the genomes we reconstructed was impressive,” Stallforth says.
Present-day bacteria act as “surrogate mothers”
All seven Chlorobium genomes were found to contain a biosynthetic gene cluster—the genetic blueprint for enzymes that produce natural products or small molecules. One particularly well-preserved Chlorobium genome was reconstructed from the roughly 19,000-year-old dental calculus retrieved from the “Red Lady of El Mirón”, the skeleton of a 35- to 40-year-old woman discovered in the El Mirón cave in northern Spain in 2010, whose name stems from the red pigment that covered her bones.
To learn what function these mysterious ancient genes had, the researchers applied state-of-the-art biotechnological tools to introduce them into living bacteria. And in point of fact: the modern-day bacteria formed functional enzymes from the ancient biosynthetic gene clusters; the enzymes in turn gave rise to two microbial natural products. While the two substances belong to the furan compound, they nevertheless constitute a new, previously unknown family. The researchers named them “paleofurans”, and they suspect the molecules played a role in regulating photosynthesis in the Chlorobium bacteria.
Pierre Stallforth says the researchers achieved two breakthroughs during the study, both of which confirmed that their idea is working: “The first was the reconstruction of ancient DNA into genomes, when we succeeded in assembling very, very large genetic sequences having more than 100,000 base pairs.” This increased the likelihood that they would find biosynthetic gene clusters. The second breakthrough was when the researchers demonstrated that the genes actually “function” in present-day bacteria.

Goal: antibiotics
“In this study, we reached a major milestone in revealing the vast genetic and chemical diversity of our microbial past,” says Christina Warinner. In future, the researchers plan to automate the processes for extracting ancient natural products, or at least embed them in standard procedures. This would allow them to discover various kinds of biosynthetic gene clusters and use computer analysis to decide which specimens should be reproduced in the lab. Their greatest hope is that some of the new natural products will be of interest to pharmaceutical companies as antibiotics.
Warinner and Stallforth stress that the breakthroughs were only possible thanks to the unique interdisciplinary collaboration in their teams: scientists from the fields of archaeology, bioinformatics, microbiology, chemistry and biotechnology all contributed to “reanimating” molecules that lived 100,000 years ago.
> The article in Science
> Project: Prehistoric medicine




