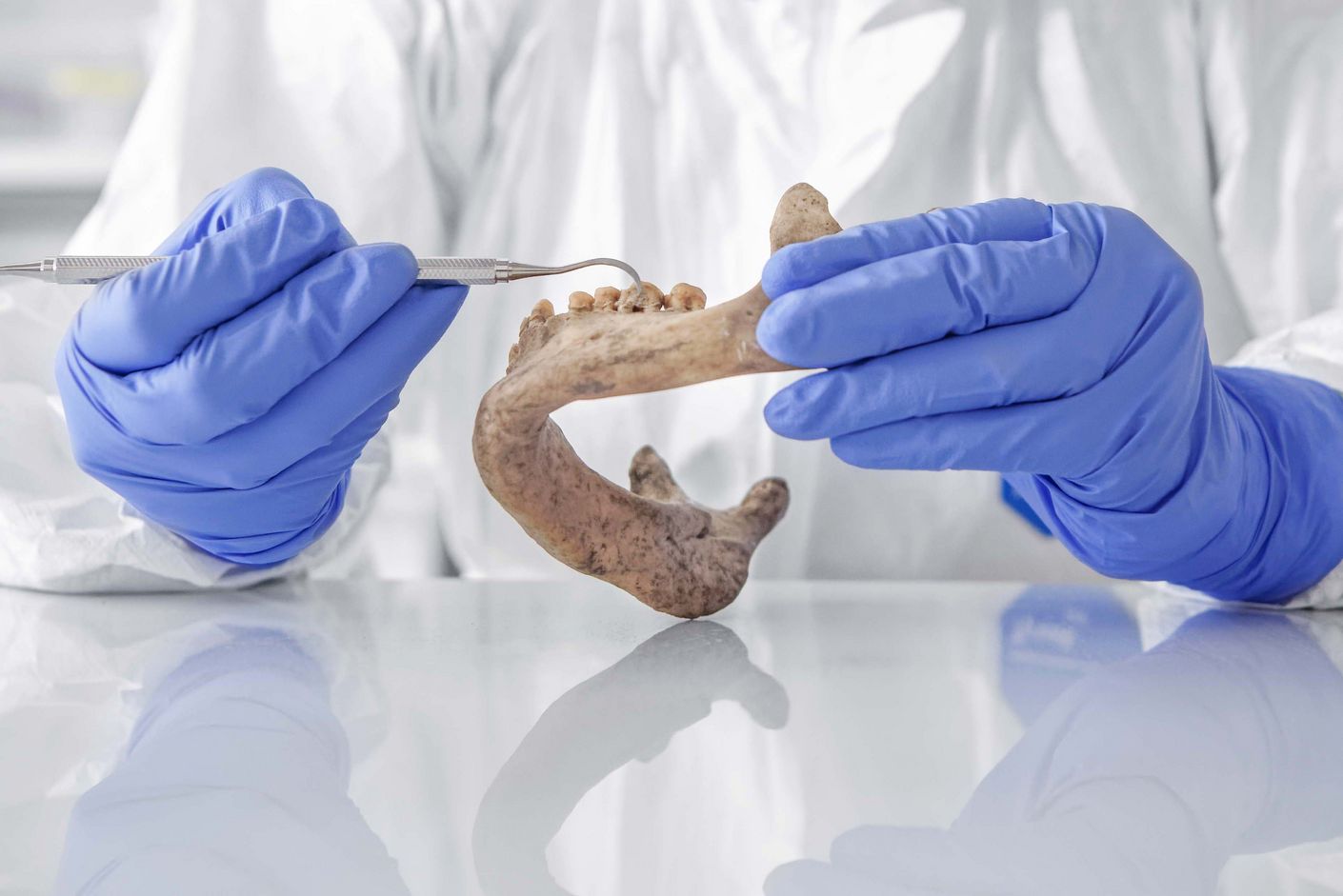
Dental plaque from down under
Since beginning their work in the new research discipline of palaeobiotechnology last year, the research team in Jena have gone far. Quite literally: their search for ancient substances with the potential to combat today’s resistant bacteria extended all the way to Oceania.
Pierre Stallforth and Christina Warinner are working together to establish palaeobiotechnology—an entirely new research discipline. The biotechnologist and archaeologist are leaders in their respective fields: Dr Christina Warinner is professor at the Max Planck Institute for the Science of Human History and at Harvard University and is specialised in the analysis of ancient DNA, while Dr Pierre Stallforth leads a research group at the Leibniz Institute for Natural Product Research and Infection Biology, where he focuses on the analysis and synthesis of natural products. With their joint project in Jena, the two researchers want to help solve an urgent problem in human medicine: resistance to antibiotics. Palaeobiotechnology offers an entirely new approach to this challenge. The researchers are delving deep into early human history in their search for substances able to combat present-day bacteria that have grown resistant to standard antibiotics. The idea in a nutshell? Warinner and Stallforth want to surprise pathogens with agents that no longer occur in nature—because today’s bacteria have had no opportunity to develop defence mechanisms against them.
Ancient DNA
The most unusual aspect of the project is that these prehistoric agents are found in the dental residue of early humans. Indeed, ancient teeth have proven a veritable gold mine for well-preserved genetic material, as both food scraps and the remains of countless bacteria are conserved in dental plaque. Because bacteria have always produced antibiotic substances—to ward off other food competitors, for example—DNA sequences responsible for producing these kinds of agents can be found in the genetic material of prehistoric bacteria. The researchers aim to identify these DNA sequences and, under laboratory conditions, insert them into the genetic material of modern bacteria. For their work, the team in Jena can draw on the thousands of archaeological specimens stored at the Max Planck Institute for the Science of Human History.
Interdisciplinary understanding
The Werner Siemens Foundation is supporting the innovative project with funding in the form of a ten-year grant. During the first year of the project, the research team managed to hold their inaugural retreat just before the coronavirus lockdown began. The aim was for the scientists in the two teams to better understand each other’s research and approach. How does an archaeologist conduct research? What does the work of a biotechnologist involve? Afterwards, the teams participated in numerous video conferences to deepen their appreciation for the “other” discipline. Project leader Pierre Stallforth says, “This work was essential in further establishing palaeobiotechnology as a new discipline.”
Sophisticated software
That the labs at the Jena research institutions were closed for two months during the coronavirus lockdown had little impact on the project’s progress. The focus for 2020 was on bioinformatics, and the researchers set to work improving existing software programs with the aim of optimising their analysis of the DNA sequences in ancient dental plaque. One of the software’s tasks is to clearly identify whether the genetic material is indeed prehistoric and not a modern-day dust speck from the excavation site. Moreover, bioinformatic data provide indications as to which snippets of genetic material from the DNA are responsible for coding antibiotic agents. The goal is to have all software tools ready by the start of 2021; that the field of bioinformatics is currently making rapid leaps thanks to artificial intelligence is of great benefit to the new discipline.
Dental plaque from around the globe
Over the course of the project’s first year, the team obtained more than 200 new dental specimens from museums and archaeological projects located in all corners of the globe. “In some cases, the genetic material is even better preserved than we had hoped,” says Christina Warinner. Specimens from Europe and Asia were already well represented in the collection, and numerous samples from Oceania were added this past year. “It’s important to have a broad range of specimens from all over the world in order to achieve maximum diversity in the bacterial DNA,” the archaeologist explains. The team’s analysis of ancient DNA also generated several publications in 2020, including a paper on the evolution of the oral microbiome over the past 100 000 years.
Grow and test
As the project progresses, the research team will identify the most promising candidates among the agents found in the dental plaque specimens. Afterwards, the next step will be to use biotechnological methodologies to insert the corresponding DNA sequences into modern-day laboratory bacteria and then grow them in a fermentation process. The antibiotic agents harvested can subsequently be tested for their effectiveness against the most common multiresistant bacteria. By 2029, the researchers aim to present interesting agents to pharmaceutical companies for the development of novel antibiotics.
Text: Adrian Ritter
Photo: Felix Wey









